European Cancer Moonshot Lund Center


We are participating in a cooperation with more than 10 countries that also involves the National Cancer Institute (NCI) in the USA. More than 75,000 patients encompassing the 15 most common cancer types will be included in this largest ever international cancer project. The purpose is to create an open database to include all types of information, e.g., personal patient information (gender, age), clinical data (results of blood samples, tissue analysis), genetic information (previous illness of relatives) etc.
_______________________________________________________________________




_______________________________________________________________________

_______________________________________________________________________
_______________________________________________________________________

_______________________________________________________________________

_______________________________________________________________________

_______________________________________________________________________

_______________________________________________________________________

_______________________________________________________________________
The European Cancer Moonshot Center has a pioneering aim to push boundaries and transcend limits in medicine by achieving what has never been done before. The main objective of the Cancer Moonshot team in Lund is to identify important diagnostic proteins that are specifically related to melanoma. The work also focuses on understanding the correlations between these proteins and the mechanisms regulated thereof.

“Regia Academia Carolina”, the main building of Lund University established in 1666

The research team in Lund
The human genome consists of approximately 20,000 protein-coding genes. If each gene codes one protein, the human proteome (the constellation of proteins expressed in different tissues and organs) would consist of 20,000 different proteins. Proteins, however, can undergo a range of processes that introduce many types of modifications. As such, the final number of proteins is completely unknown. Anything from 20,000 to several millions has been reported in the literature (1,2). Amongst all these proteins, the Cancer Moonshot Lund chapter will attempt to identify those that are specifically related to melanoma.

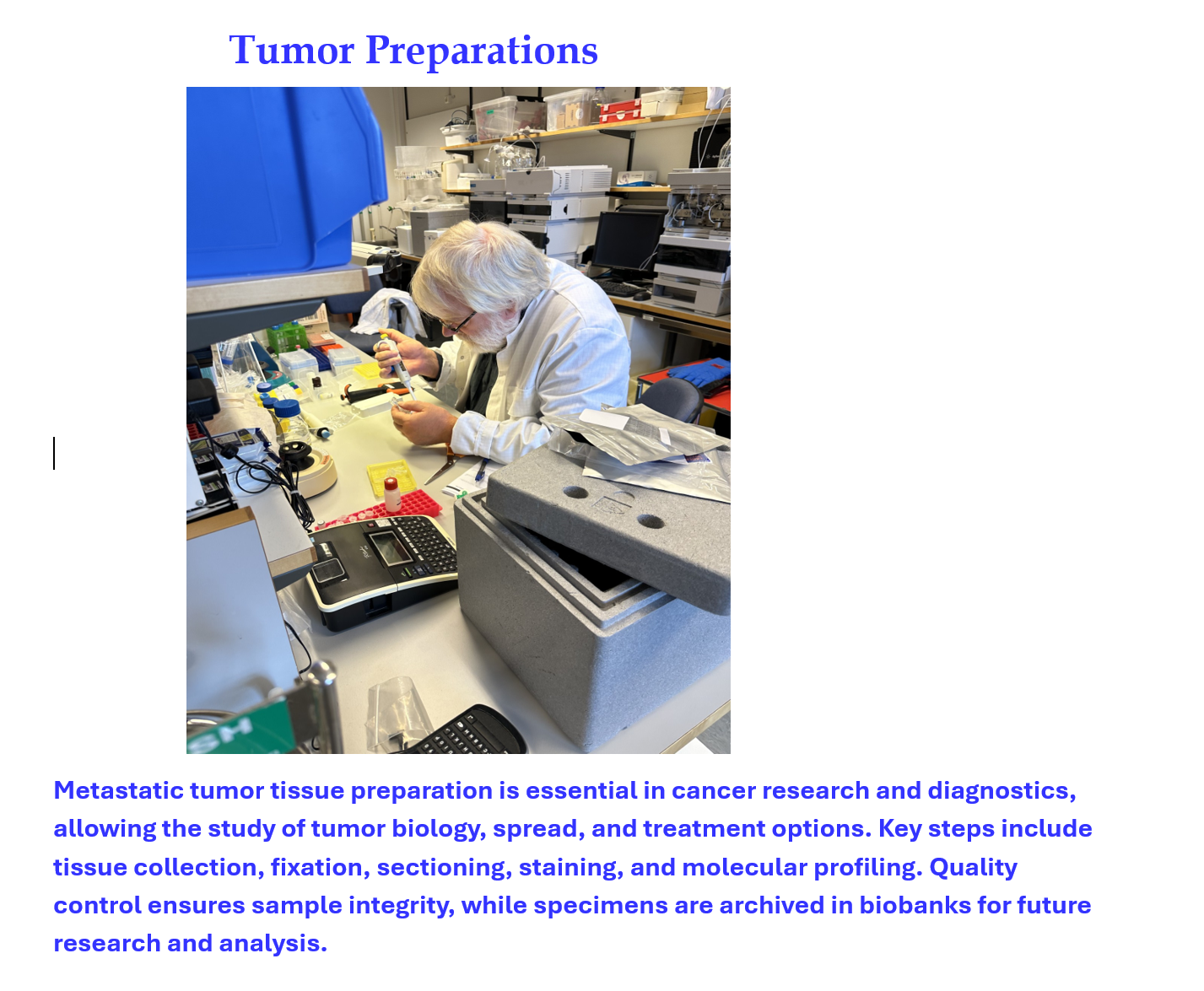
One of the preparation laboratories
Depending on the progress of the disease and the treatment used, these proteins can alter in quantity. Some proteins increase (upregulated, higher concentration) and some decrease in quantity (downregulated, lower concentration). The task is to determine the pattern and to understand the pathways involved in such a complex system. Eventually, identification of the most significant indicative proteins will then aid in the diagnosis and treatment of cancer patients.
When these proteins are identified it will be easier to determine if there is indeed melanoma, what type of melanoma, and how the cancer should be treated in the most efficient way; so-called target-driven treatment.
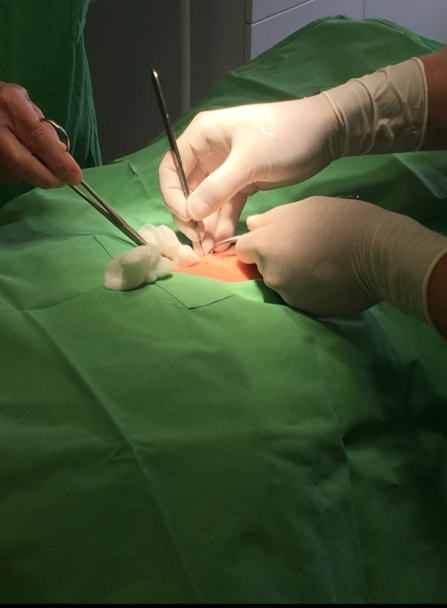
Within our clinical activities, we have encountered compelling patient cases that provide valuable insights into the dynamic progression of diseases. These instances involve a meticulous tracking of the journey from primary tumors to subsequent relapses following various treatments. What makes these cases particularly noteworthy is the rapid emergence of satellite metastasis, showcasing the intricacies of disease evolution.
As we delve into the details of these patient narratives, we observe the complexities involved in understanding how tumors respond to treatments and the subsequent development of relapses. The cases serve as a lens into the challenges faced in clinical settings, shedding light on the unpredictable nature of disease trajectories.
In these patient journeys, the swift onset of satellite metastasis becomes a focal point of exploration. This phenomenon not only underscores the aggressiveness of certain diseases but also presents a formidable challenge in the treatment landscape. The progression from a manageable stage to an advanced and ultimately incurable disease stage emphasizes the urgency and importance of developing novel strategies for intervention and management.
By closely examining these patient cases, we aim to enhance our understanding of the underlying mechanisms driving disease progression, contributing valuable data to inform future research and the development of more effective treatment approaches.


Our centre offers a protein sequencing laboratory and a fully automated biobank. The procedure is described in the flowchart below:
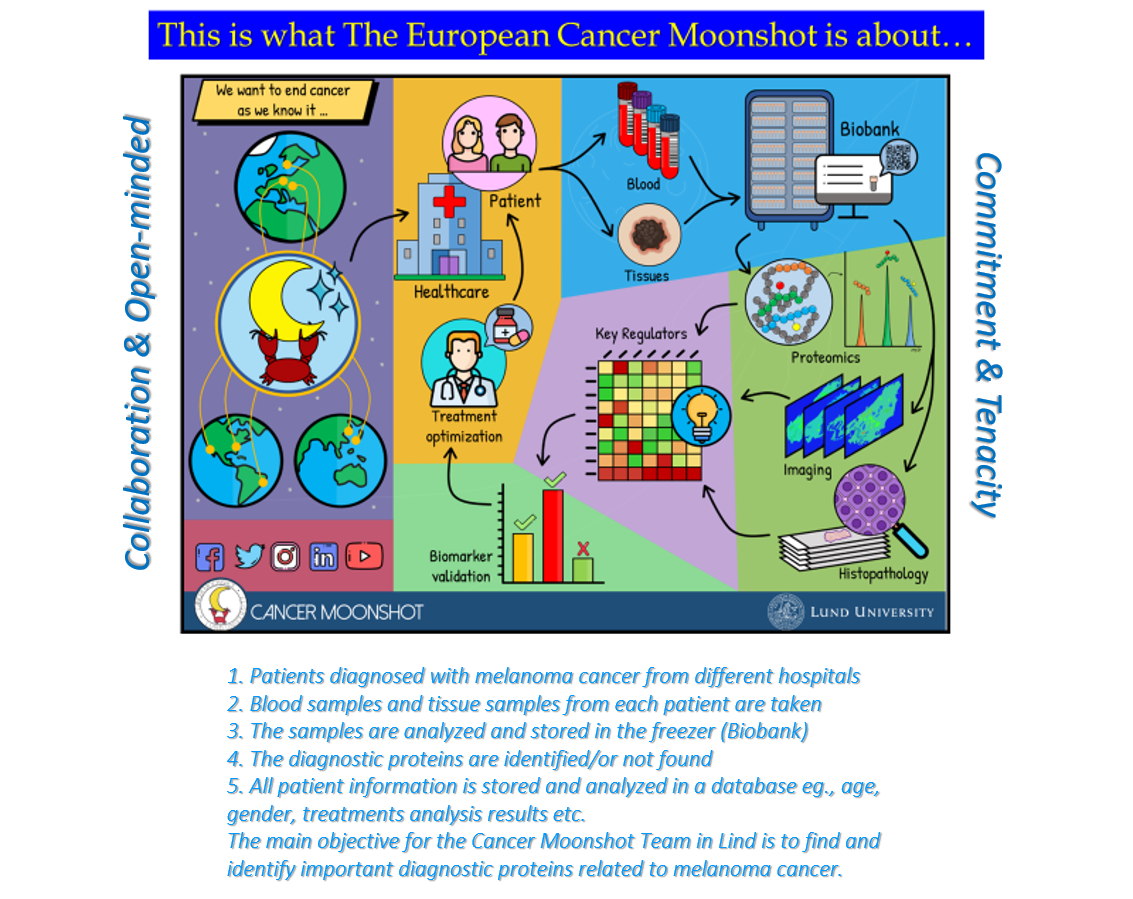
- Blood and tissue collection from melanoma patients.
- Sample storage at -80°C in large robotic-controlled freezers in the biobank.
- Analysis of the samples by the latest-generation high-resolution mass spectrometry to identify proteins.
- Bioinformatics: powerful statistical software to correlate identified proteins with clinical data and presentation of the data in a simple format.
___
REFERENCES
1. Elena A. Ponomarenko, Ekaterina V. Poverennaya, Ekaterina V. Ilgisonis, et al., “The Size of the Human Proteome: The Width and Depth”,
International Journal of Analytical Chemistry, May 19, 2016, Article ID 7436849.
2. Ruedi Aebersold1 , Jeffrey N Agar2 , I Jonathan Amster, et al., “How many human proteoforms are there?”,
Nature Chemical Biology, Feb 14;14(3):206-214 (2018).